Best practices for multi-sample variant calling with DeepVariant (WES trio demonstration)
Best practices for multi-sample variant calling with DeepVariant (WES trio demonstration)
Overview
This document outlines all the steps and considerations for calling and merging a trio using DeepVariant and GLnexus. These best practices were developed and evaluated as described in the article published in Bioinformatics: Accurate, scalable cohort variant calls using DeepVariant and GLnexus (2021).
The process involves 3 major stages: running DeepVariant to create individual genome call sets, running GLnexus to merge call sets, and analyzing the merged call set.
NOTE: This case study demonstrates an example of how to run DeepVariant end-to-end on one machine. The steps below were done on a machine with this example command to start a machine.
The steps in this document can be extended to merge larger cohorts as well.
See this workflow:
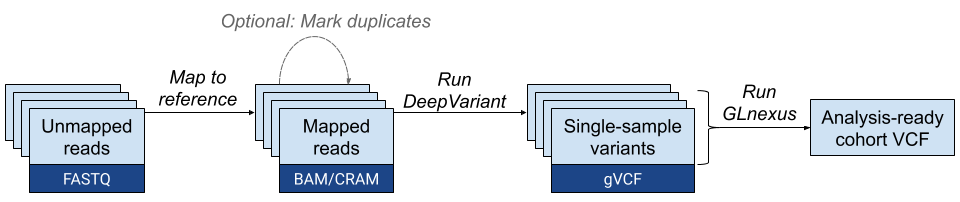
A few things to note before we start:
- It is recommended to use BAM files with original quality scores. In the case
that BAM files went through recalibration, optional DV flags can be used in
order to use original scores:
--parse_sam_aux_fields,--use_original_quality_scores. - DeepVariant optionally allows gVCF output. This option is required for further GLnexus analysis in this document.
Dataset
The Whole Exome Sequencing (WES) dataset we’re using is from:
ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/
- HG002_NA24385_son
- HG003_NA24149_father
- HG004_NA24143_mother
Commands for downloading the input BAMs
Just for convenience, we use aria2 to download our data. You can change it to whatever other tools (wget, curl) that you prefer.
To install aria2, you can run: sudo apt-get -y install aria2
DIR="${PWD}/trio"
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/OsloUniversityHospital_Exome/151002_7001448_0359_AC7F6GANXX_Sample_HG002-EEogPU_v02-KIT-Av5_AGATGTAC_L008.posiSrt.markDup.bam -o HG002.bam
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/OsloUniversityHospital_Exome/151002_7001448_0359_AC7F6GANXX_Sample_HG002-EEogPU_v02-KIT-Av5_AGATGTAC_L008.posiSrt.markDup.bai -o HG002.bai
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG003_NA24149_father/OsloUniversityHospital_Exome/151002_7001448_0359_AC7F6GANXX_Sample_HG003-EEogPU_v02-KIT-Av5_TCTTCACA_L008.posiSrt.markDup.bam -o HG003.bam
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG003_NA24149_father/OsloUniversityHospital_Exome/151002_7001448_0359_AC7F6GANXX_Sample_HG003-EEogPU_v02-KIT-Av5_TCTTCACA_L008.posiSrt.markDup.bai -o HG003.bai
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG004_NA24143_mother/OsloUniversityHospital_Exome/151002_7001448_0359_AC7F6GANXX_Sample_HG004-EEogPU_v02-KIT-Av5_CCGAAGTA_L008.posiSrt.markDup.bam -o HG004.bam
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG004_NA24143_mother/OsloUniversityHospital_Exome/151002_7001448_0359_AC7F6GANXX_Sample_HG004-EEogPU_v02-KIT-Av5_CCGAAGTA_L008.posiSrt.markDup.bai -o HG004.bai
Command for downloading the reference file
aria2c -c -x10 -s10 -d "${DIR}" https://storage.googleapis.com/deepvariant/exome-case-study-testdata/hs37d5.fa.gz
gunzip ${DIR}/hs37d5.fa.gz
aria2c -c -x10 -s10 -d "${DIR}" https://storage.googleapis.com/deepvariant/exome-case-study-testdata/hs37d5.fa.fai
Command for downloading the input capture region BED file
aria2c -c -x10 -s10 -d "${DIR}" https://storage.googleapis.com/deepvariant/exome-case-study-testdata/agilent_sureselect_human_all_exon_v5_b37_targets.bed
Command for downloading the truth files
HG002:
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio/HG002_NA24385_son/NISTv4.2.1/GRCh37/HG002_GRCh37_1_22_v4.2.1_benchmark.vcf.gz -o HG002_truth.vcf.gz
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio/HG002_NA24385_son/NISTv4.2.1/GRCh37/HG002_GRCh37_1_22_v4.2.1_benchmark.vcf.gz.tbi -o HG002_truth.vcf.gz.tbi
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio/HG002_NA24385_son/NISTv4.2.1/GRCh37/HG002_GRCh37_1_22_v4.2.1_benchmark_noinconsistent.bed -o HG002_truth.bed
HG003:
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio/HG003_NA24149_father/NISTv4.2.1/GRCh37/HG003_GRCh37_1_22_v4.2.1_benchmark.vcf.gz -o HG003_truth.vcf.gz
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio/HG003_NA24149_father/NISTv4.2.1/GRCh37/HG003_GRCh37_1_22_v4.2.1_benchmark.vcf.gz.tbi -o HG003_truth.vcf.gz.tbi
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio/HG003_NA24149_father/NISTv4.2.1/GRCh37/HG003_GRCh37_1_22_v4.2.1_benchmark_noinconsistent.bed -o HG003_truth.bed
HG004:
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio/HG004_NA24143_mother/NISTv4.2.1/GRCh37/HG004_GRCh37_1_22_v4.2.1_benchmark.vcf.gz -o HG004_truth.vcf.gz
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio/HG004_NA24143_mother/NISTv4.2.1/GRCh37/HG004_GRCh37_1_22_v4.2.1_benchmark.vcf.gz.tbi -o HG004_truth.vcf.gz.tbi
aria2c -c -x10 -s10 -d "${DIR}" ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio/HG004_NA24143_mother/NISTv4.2.1/GRCh37/HG004_GRCh37_1_22_v4.2.1_benchmark_noinconsistent.bed -o HG004_truth.bed
(No need to install bcftools and other tools, because they are now installed in the DeepVariant images.)
Run DeepVariant on trio to get 3 single sample VCFs
First, install docker if you don’t have it yet: sudo apt-get -y install
docker.io
With the example command below, it runs DeepVariant on the trio one by one. This is for demonstration only. If you’re running this on a large cohort, running serially is not the most effective approach.
N_SHARDS=$(nproc) # Or change to the number of cores you want to use
CAPTURE_BED=agilent_sureselect_human_all_exon_v5_b37_targets.bed
VERSION=1.5.0
declare -a trio=(HG002 HG003 HG004)
for SAMPLE in "${trio[@]}"
do
BAM=${SAMPLE}.bam
OUTPUT_VCF=${SAMPLE}.vcf.gz
OUTPUT_GVCF=${SAMPLE}.g.vcf.gz
time sudo docker run \
-v "${DIR}":"/data" \
google/deepvariant:${VERSION} \
/opt/deepvariant/bin/run_deepvariant \
--model_type=WES \
--ref="/data/hs37d5.fa" \
--reads="/data/${BAM}" \
--regions="/data/${CAPTURE_BED}" \
--output_vcf="/data/${OUTPUT_VCF}" \
--output_gvcf="/data/${OUTPUT_GVCF}" \
--num_shards=${N_SHARDS}
done
Note: The BAM files should provide unique names for each sample in their SM
header tag, which is usually derived from a command-line flag to the read
aligner. If your BAM files don’t have unique SM tags (and if it’s not feasible
to adjust the alignment pipeline), add the --sample_name=XYZ flag to
run_deepvariant to override the sample name written into the gVCF file header.
Merge the trio samples using GLnexus
Run GLnexus to merge 3 gVCFs
And then run GLnexus with this config:
sudo docker pull quay.io/mlin/glnexus:v1.2.7
time sudo docker run \
-v "${DIR}":"/data" \
quay.io/mlin/glnexus:v1.2.7 \
/usr/local/bin/glnexus_cli \
--config DeepVariantWES \
--bed "/data/${CAPTURE_BED}" \
/data/HG004.g.vcf.gz /data/HG003.g.vcf.gz /data/HG002.g.vcf.gz \
| sudo docker run -i google/deepvariant:${VERSION} bcftools view - \
| sudo docker run -i google/deepvariant:${VERSION} bgzip -c \
> ${DIR}/deepvariant.cohort.vcf.gz
When we ran on this WES trio, it took only about 13 seconds. For more details on performance, see GLnexus performance guide.
For a WGS cohort, we recommend using --config DeepVariantWGS instead of
DeepVariantWES. Another preset DeepVariant_unfiltered is available in
glnexus:v1.2.7 or later versions for merging DeepVariant gVCFs with no QC
filters or genotype revision (see
GitHub issue #326 for a
potential use case). The details of these presets can be found
here.
Annotate the merged VCF with Mendelian discordance information using RTG Tools
Create an SDF template from our reference file:
sudo docker run \
-v "${DIR}":"/data" \
realtimegenomics/rtg-tools format \
-o /data/hs37d5.sdf /data/hs37d5.fa
Create a PED file $DIR/trio.ped that looks like this (with the sample name
of the trio):
#PED format pedigree
#
#fam-id/ind-id/pat-id/mat-id: 0=unknown
#sex: 1=male; 2=female; 0=unknown
#phenotype: -9=missing, 0=missing; 1=unaffected; 2=affected
#
#fam-id ind-id pat-id mat-id sex phen
1 Sample_Diag-excap51-HG002-EEogPU Sample_Diag-excap51-HG003-EEogPU Sample_Diag-excap51-HG004-EEogPU 1 0
1 Sample_Diag-excap51-HG003-EEogPU 0 0 1 0
1 Sample_Diag-excap51-HG004-EEogPU 0 0 2 0
Annotate merged VCF with RTG Tools
sudo docker run \
-v "${DIR}":"/data" \
realtimegenomics/rtg-tools mendelian \
-i /data/deepvariant.cohort.vcf.gz \
-o /data/deepvariant.annotated.vcf.gz \
--pedigree=/data/trio.ped \
-t /data/hs37d5.sdf \
| tee ${DIR}/deepvariant.input_rtg_output.txt
The output is:
Checking: /data/deepvariant.cohort.vcf.gz
Family: [Sample_Diag-excap51-HG003-EEogPU + Sample_Diag-excap51-HG004-EEogPU] -> [Sample_Diag-excap51-HG002-EEogPU]
Concordance Sample_Diag-excap51-HG002-EEogPU: F:58220/58735 (99.12%) M:58627/58749 (99.79%) F+M:58003/58640 (98.91%)
Sample Sample_Diag-excap51-HG002-EEogPU has less than 99.0 concordance with both parents. Check for incorrect pedigree or sample mislabelling.
827/58974 (1.40%) records did not conform to expected call ploidy
58870/58974 (99.82%) records were variant in at least 1 family member and checked for Mendelian constraints
189/58870 (0.32%) records had indeterminate consistency status due to incomplete calls
649/58870 (1.10%) records contained a violation of Mendelian constraints
From this report, we know that there is a 1.10% Mendelian violation rate, and
0.32% of the records had incomplete calls (with .) so RTG couldn’t determine
whether there is violation or not.
Single sample quality metrics
In addition to the cohort quality statistics, for completeness we generate single-sample quality metrics.
ti/tv ratio
We run bcftools stats on the 3 VCF outputs. Since our DeepVariant run already
constrained to just the capture regions, no need to specify it again here. We
had to pass in the -f PASS flag so that only the PASS calls are considered.
declare -a trio=(HG002 HG003 HG004)
for SAMPLE in "${trio[@]}"
do
sudo docker run \
-v ${DIR}:${DIR} \
google/deepvariant:${VERSION} \
bcftools stats -f PASS \
${DIR}/${SAMPLE}.vcf.gz \
> ${DIR}/${SAMPLE}.stats
done
| Sample | [3]ts | [4]tv | [5]ts/tv | [6]ts (1st ALT) | [7]tv (1st ALT) | [8]ts/tv (1st ALT) |
|---|---|---|---|---|---|---|
| HG002 | 29884 | 11664 | 2.56 | 29871 | 11644 | 2.57 |
| HG003 | 29760 | 11718 | 2.54 | 29750 | 11700 | 2.54 |
| HG004 | 30003 | 11816 | 2.54 | 29991 | 11799 | 2.54 |
If you want to restrict to the truth BED files, use this command:
declare -a trio=(HG002 HG003 HG004)
for SAMPLE in "${trio[@]}"
do
sudo docker run \
-v ${DIR}:${DIR} \
google/deepvariant:${VERSION} \
bcftools stats -f PASS \
-T ${DIR}/${SAMPLE}_truth.bed \
${DIR}/${SAMPLE}.vcf.gz \
> ${DIR}/${SAMPLE}.with_truth_bed.stats
done
Which resulted in this table:
| Sample | [3]ts | [4]tv | [5]ts/tv | [6]ts (1st ALT) | [7]tv (1st ALT) | [8]ts/tv (1st ALT) |
|---|---|---|---|---|---|---|
| HG002 | 27706 | 10538 | 2.63 | 27698 | 10525 | 2.63 |
| HG003 | 27335 | 10513 | 2.60 | 27331 | 10503 | 2.60 |
| HG004 | 27485 | 10601 | 2.59 | 27478 | 10590 | 2.59 |
Rtg vcfstats
declare -a trio=(HG002 HG003 HG004)
for SAMPLE in "${trio[@]}"
do
sudo docker run \
-v "${DIR}":"/data" \
realtimegenomics/rtg-tools vcfstats \
/data/${SAMPLE}.vcf.gz \
> ${DIR}/${SAMPLE}.vcfstats
done
which shows the following:
HG002:
Location : /data/HG002.vcf.gz
Failed Filters : 14706
Passed Filters : 45150
SNPs : 41515
MNPs : 0
Insertions : 1854
Deletions : 1755
Indels : 22
Same as reference : 4
SNP Transitions/Transversions: 2.56 (41767/16309)
Total Het/Hom ratio : 1.49 (27009/18137)
SNP Het/Hom ratio : 1.51 (24977/16538)
MNP Het/Hom ratio : - (0/0)
Insertion Het/Hom ratio : 1.06 (954/900)
Deletion Het/Hom ratio : 1.51 (1056/699)
Indel Het/Hom ratio : - (22/0)
Insertion/Deletion ratio : 1.06 (1854/1755)
Indel/SNP+MNP ratio : 0.09 (3631/41515)
HG003:
Location : /data/HG003.vcf.gz
Failed Filters : 15562
Passed Filters : 45011
SNPs : 41448
MNPs : 0
Insertions : 1829
Deletions : 1714
Indels : 19
Same as reference : 1
SNP Transitions/Transversions: 2.52 (41586/16521)
Total Het/Hom ratio : 1.47 (26806/18204)
SNP Het/Hom ratio : 1.49 (24810/16638)
MNP Het/Hom ratio : - (0/0)
Insertion Het/Hom ratio : 1.08 (951/878)
Deletion Het/Hom ratio : 1.49 (1026/688)
Indel Het/Hom ratio : - (19/0)
Insertion/Deletion ratio : 1.07 (1829/1714)
Indel/SNP+MNP ratio : 0.09 (3562/41448)
HG004:
Location : /data/HG004.vcf.gz
Failed Filters : 15281
Passed Filters : 45400
SNPs : 41786
MNPs : 0
Insertions : 1856
Deletions : 1735
Indels : 18
Same as reference : 5
SNP Transitions/Transversions: 2.55 (41616/16328)
Total Het/Hom ratio : 1.57 (27705/17690)
SNP Het/Hom ratio : 1.59 (25649/16137)
MNP Het/Hom ratio : - (0/0)
Insertion Het/Hom ratio : 1.13 (983/873)
Deletion Het/Hom ratio : 1.55 (1055/680)
Indel Het/Hom ratio : - (18/0)
Insertion/Deletion ratio : 1.07 (1856/1735)
Indel/SNP+MNP ratio : 0.09 (3609/41786)
Run hap.py to calculate the accuracy of DeepVariant generated call sets
sudo docker pull jmcdani20/hap.py:v0.3.12
declare -a trio=(HG002 HG003 HG004)
for SAMPLE in "${trio[@]}"
do
sudo docker run -i \
-v "${DIR}":"/data" \
jmcdani20/hap.py:v0.3.12 /opt/hap.py/bin/hap.py \
"/data/${SAMPLE}_truth.vcf.gz" \
"/data/${SAMPLE}.vcf.gz" \
-f "/data/${SAMPLE}_truth.bed" \
-T "/data/${CAPTURE_BED}" \
-r "/data/hs37d5.fa" \
-o "/data/${SAMPLE}.happy.output" \
--engine=vcfeval \
--pass-only > ${DIR}/${SAMPLE}.stdout
done
Accuracy F1 scores:
| Sample | Indel | SNP |
|---|---|---|
| HG002 | 0.976009 | 0.993962 |
| HG003 | 0.967468 | 0.993909 |
| HG004 | 0.973334 | 0.994004 |